Does evolution estimate gradients?
Visual summary on Twitter
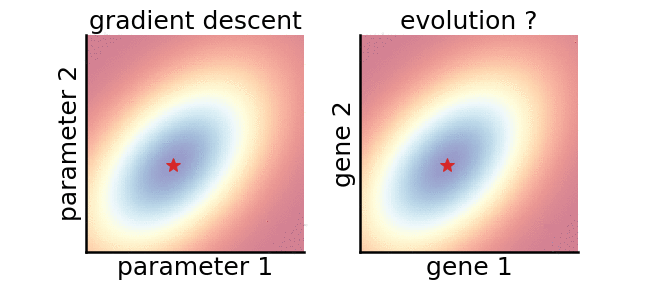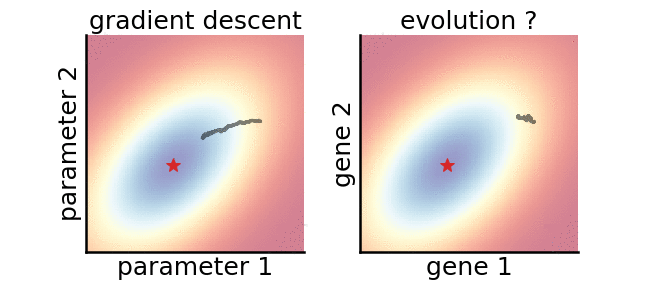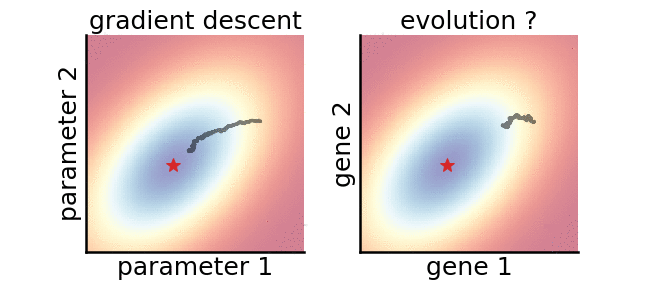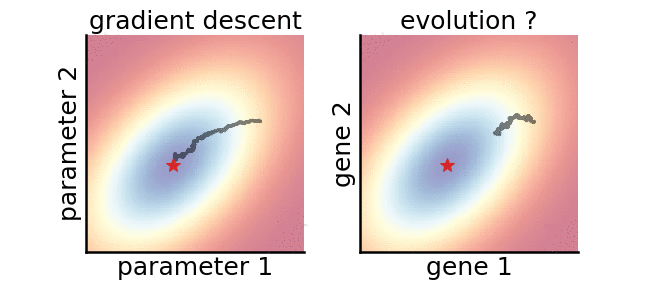
A gradient-based (left) and a gradient-free (right) algorithm that minimise the same noisy quadratic loss function. Color indicates performance.
Imagine you’re on a hike, trying to work your way up a mountain. Unfortunately, it’s a foggy day, so you can see your immediate surroundings but not the mountain top. In which direction do you decide to walk? Your intuition probably tells you to choose the direction in which the terrain around you rises most rapidly. And this intuition makes sense: Without knowledge about the overall landscape, the immediate direction of steepest ascent is the most promising1.
The same intuition underlies many optimisation algorithms, including those that power deep neural networks. Instead of changing hiking directions, these algorithms change network parameters, and instead of getting up a mountain, they improve the network’s performance. But the basic idea is the same: Make a change in the direction of steepest improvement.
Mathematically, the direction of steepest improvement is known as the gradient of the objective function, and the overall approach is known as gradient ascent (or descent). Optimization algorithms that don’t use gradients are typically less sample efficient than algorithms that do, and deep learning would not enjoy its current success without them2.
Yet, evolution by natural selection — arguably the most impressive optimisation algorithm we know — does not exploit this powerful principle. At least it doesn’t according to conventional wisdom, which says that evolution relies on random genetic changes (mutations). Futuyma and Kirkpatrick write in their authoritative textbook Evolution:
Mutations are random with respect to what will improve survival and reproduction. New conditions do not increase the frequency of mutations that are beneficial in those conditions.
The only non-random part of evolution, the part responsible for improvements in fitness, is therefore natural selection acting upon mutations — not the mutations themselves. This can come as a surprise to anyone familiar with gradient descent and its benefits. Even gradient-free algorithms inspired by evolution often sample “mutations” based on gradient estimates. How, then, could biological evolution produce its astonishing results based on random mutations?
This question touches upon the fundamental mechanisms of evolution: What explains biological diversity, and what explains the often exquisite adaptation of this diversity to an organism’s needs? For example: The different beaks of Darwin’s finches, each adapted to local food sources — are they just attributable to selection, or also to well-targeted mutations?
In this blog post I investigate why textbooks teach that evolution works through random mutations, and to what extent this might change in the light of recent experiments. Let’s start by determining what a realistic experimental test of non-random mutations would look like.
Testing for non-random mutations is hard
Experimental evidence for non-random mutations would need to show that (1) certain genes mutate more than others, and (2) this difference in mutation rates is adaptive, i.e. increases fitness. It might be a stretch to conclude from this that “evolution estimates gradients”, but adaptive variability in mutation rates would at least imply that mutations are more aligned with the gradient than expected by chance.
Many studies have indeed found less variability in some genes than in others. Unfortunately, this variability is the combined outcome of mutation and selection, and therefore doesn’t necessarily arise from variability in mutation rates. A lack of variability in a certain gene, for example, doesn’t mean the gene never mutates. Instead, it could mutate just as often as other genes, but whenever it does, the mutation is eliminated by purifying selection.
To find non-random mutations, we therefore need to catch mutagenesis in the act, detecting mutations as they happen. This is hard, because mutations are rare3, and even if we did find certain mutations to occur more often than others — we still would not know their fitness effect, and wouldn’t be able to tell if the mutational bias was adaptive. An experimental test for non-random mutations experiment should therefore use a fast-growing model organism to allow for sufficient mutations, and a mutation with clear fitness effects to test for adaptation.
Bacteria lead the way

Salvador Luria (source), bacteriophages attacking an E. coli bacterium (source).
The biologist Salvador Luria would be the first to crack this challenge. Luria chose the bacterium Escherichia coli as a model organism. Most of his colleagues did not consider lowly bacteria relevant for answering deep evolutionary problems. But Luria, a relative newcomer in the field, suspected something we now know to be true: Many molecular mechanisms of evolution are shared across the tree of life, all the way from humans to bacteria. E. coli’s 20 minute replication cycle allowed Luria to go from a single bacterium to millions in less than a day. Enough replication cycles for many mutations to occur.
Luria had also determined an E. coli mutation with a clear fitness effect. Like other bacteria, E. coli is involved in an intense arms race with bacteriophages (viruses that infect bacteria). One such virus is the T1 phage, which kills E. coli bacteria. But some bacteria are resistant to T1, because of a mutation in the receptor protein to which T1 attaches itself. Luria reasonsed that this mutation determined life or death during a T1 invasion — a clear fitness readout.
Luria decided to investigate if E. coli’s T1 resistance occurred spontaneously, or in response to the T1 threat. Only the second outcome would be consistent with non-random and adaptive mutations. But a key challenge still remained: how to tell the two hypotheses apart?
A Nobel-prize winning experiment

The Luria-Delbruck experiment, for which the competing hypotheses of induced and spontaneous mutations predict different numbers of surviving bacteria. Source: Wikipedia.
First, suppose resistance was induced by the presence of T1. If we expose many equally sized E. coli populations to T1, the law of large numbers predicts that each population has similar numbers of survivors.
The spontaneous resistance model, on the other hand, predicts that the number of survivors should fluctuate across populations. If mutations occurred absent T1, this could have happened any time during the growth of the population. Had it occurred at the very start, most of the population should have inherited the resistance, and would have survived. But had it occurred at the very end, only a few members would have survived.
So here’s the experiment Luria conducted. He let individual bacteria replicate until each had generated a large population. Thanks to E. coli’s fast replication times, this took only a day. Then, he added T1 phages to the populations, invariably killing. many bacteria. Some survived, though, and the number of survivors varied by several orders of magnitude across different populations. In those with many survivors, T1 resistance must have occurred long before the T1 exposure, rather than in response to the T1 presence.
Their findings earned Luria (along with Max Delbruck and Alfred Hershey) the 1969 Nobel prize in Physiology or Medicine. By then, other experiments had supported their conclusion that mutations occur independently of their fitness effects, establishing it as a central principle of evolutionary genetics4.
But like the original work from Luria and Delbruck, the follow-up reports of random mutations were held back by a major technological limitation: They couldn’t detect mutations across multiple genes. At the time of their key experiment, Luria and Delbruck didn’t even know what a gene exactly was! This meant only those “genes” could be studied that had a clear fitness effect. But most genes, we know now, affect fitness in only a modest way. The unbiased study of mutation rates therefore had to wait until the post-genomic era.
Genomic clues of non-random mutation rates
In 2012, Martincorena et al. conducted such an unbiased test by comparing 34 E. coli genomes. They couldn’t simply compare the diversity across genes, since this is shaped by both mutations and selection. The authors therefore focused on the so-called synonymous diversity between the same genes in different E. coli strains, and statistically accounted for factors other than mutation rate. Whereas genes with smaller diversity across organisms might simply experience stronger purifying selection, genes with smaller synonymous diversity are actually thought to have a smaller mutation rate.

(a) Mutation rates (estimated using the synonymous diversity across different strains) vary along the E. coli genome. Red bars indicate the 95% confidence interval under a uniform mutation rate. MB: megabases, or 1000 base pairs. (b) Genes with lower mutation rates are often essential. From Martincorena et al..
The authors found the synonymous diversity to vary 20-fold across the E. coli genome, with certain genes showing much less diversity than expected from a uniform mutation rate, and others showing much more (figure, panel a). This pattern correlated with function: Diversity was smallest for essential genes (those critical for healthy function) and those under strong purifying selection (panel b). Martincorena et al. therefore concluded that the mutation rate varies across the genome, and that this might be a “risk management strategy” to minimise the probability of bad mutations.
The finding made a splash in the world of evolutionary biology, but soon other biologists started to point at potential problems (Chen & Zhang, Maddamsetti et al.). One problem was theoretical: Mutations to individual genes occur so rarely that the advantage of an even lower and gene-specific mutation rate is too small to evolve (more on this later). Other challenges were empirical in nature. For example, Martincorena et al. aimed to eliminate the effect of selection by analysing only synonymous genetic diversity, but might have failed to account for all confounding factors.
Definitive evidence for biased mutations therefore required a stronger, experimental rather than statistical, control for selection.
Planting the seeds of a revolution
A rigorous but time-consuming approach to experimentally minimise the effect of selection is a so-called mutation accumulation experiment in which one randomly selects the organism that will give rise to the next generation5. Random selection rules out natural selection by making reproductive success independent of fitness. Repeating the process for multiple generations results in the accumulation of mutations untouched by selection; repeating it in parallel reveals if certain mutations arise more frequently than others (see figure).

A typical mutation accumulation (MA) experiment, in which n lines are bred for t generations. In the main dataset from Monroe et al., $n=107$ and $t = 25$. Source: Halligan & Keightley.
Recently, Grey Monroe et al. used mutation accumulation lines to investigate the randomness of mutations in Arabidopsis thaliana (the primary model for plant genetics, see figure a). The authors found that genetic diversity across mutation accumulation lines greatly varied across the genome, being reduced by a half in gene bodies (figure b). This time around, the reduced variability really could not be due to selection, and therefore had to be attributed to variability in mutation rates.

(a): Arabidopsis thaliana. (b): mutations are lower for bases within gene bodies. TSS: transcription start sites, TTS: transcription termination sites. (c): the mutation rate varies with gene function. Adapted from Monroe et al..
Monroe et al. also investigated potential mechanisms, building on ideas from Martincorena & Luscombe and others. The authors showed that the mutation rate of a genomic region can be predicted from several of its features. Some of these are epigenomic, i.e. they affect gene expression without changing the DNA itself6, for example by changing the histone proteins that package DNA. This change in packaging can increase or decrease the genes’ expression by making it more or less accessible to transcription factors. Interestingly, such a change in a gene’s packaging can also increase the accuracy of its replication (see e.g. Chong et al.). Natural selection could thus tune mutation rates by shaping DNA packaging around important genes.
Finally, the authors also provide evidence that the mutation rate variability is adaptive: Essential genes mutate less than genes with an environmentally-dependent function (Fig c). The variation in mutation rates therefore reduces the number of deleterious mutations, allowing it to bias the course of evolution.
Reconciling the sequencer and the petri dish
The genomic data from Martincorena and Monroe provide strong evidence that natural selection can tune local mutation rates. So why didn’t Luria and Delbruck find that T1 presence increased the likelihood of mutations when this clearly would have been adaptive?
First, Luria & Delbruck tested for an increased mutation rate in a gene under positive selection. The genomic data, by contrast, provide evidence for decreased mutation rates in genes under negative selection. In fact, Martincorena specifically looked for — but failed to find — increased mutation rates in genes under positive selection. The difference between decreasing the mutation rate of some genes and increasing the rate of others is not merely semantic. Increased mutation rates, on one hand, would indicate selection for more change in the right direction. But the current data don’t strongly support this idea. Instead, they indicate that natural selection mainly tries to prevent rather than elicit change7.
The second difference between classical and recent experiments is the length of the genomic region across which mutation rates might vary. Luria & Delbruck found that the mutation rate of a single gene (that for a receptor protein) did not increase under positive selection. Martincorena & Monroe, on the other hand, found that mutation rates vary across many genes at once. One of the key epigenetic marks that could decrease mutation rates is shared by about 15% of the arabidopsis genome.
Tuning the mutation rate of many genes at once
Both Luria & Delbruck and the genomic data are therefore consistent with the idea that mutation rates vary across the genome, but not on a per-gene basis. This bolsters the case for non-random mutation rates by addressing the earlier concern that selection is too weak to tune mutation rates of individual genes.
In general, selection is strong enough to establish a trait if its fitness advantage overcomes other evolutionary forces8, in particular genetic drift (evolution by chance). We need:
advantage > genetic drift
Genetic drift is stronger in small populations, just like chance plays a larger role in determining the average outcome of, say, 10 versus 10000 coin tosses. Specifically, the strength of genetic drift is assumed to be proportional to the inverse population size9:
Genetic drift = 1/population size.
The expected fitness advantage, on the other hand, can be decomposed into three terms:
Advantage = mutation rate * change in mutation rate * effect of mutation.
Selection is therefore strong enough to tune the mutation rate of a particular gene if
mutation rate * change in mutation rate * effect of mutation > 1/population size.
As we’ve seen, mutation rates are extremely low, and the fitness effect of most mutations is small. The inequality is therefore not satisfied—selection on a gene-specific mutation rate is overwhelmed by genetic drift. But this changes when considering tuning mutation rates of many genes at once (Martincorena & Luscombe). The fitness advantage scales with the size L of the genomic segment that is affected together:
mutation rate * change in mutation rate * effect of mutation * L > 1/population size.
Monroe et al. do the maths and show that, in arabidopsis, the minimum length L is much smaller than the 15% of the genome affected by the mechanisms thought to tune mutation rates. These mechanisms therefore could have been shaped by selection.
Conclusion
In sum, recent data suggests that mutations do not arise independently of their fitness effect. This is, I think, quite revolutionary, and deserves to be added to the textbooks.
Although the mutational bias seems adaptive, it does not warrant the conclusion that evolution “estimates the fitness gradient”. What would? First, evidence that selection can not just selectively decrease but also increase mutation rates. Second, that it does so in a more specific way than across, say, 15% of the genome.
Awaiting future experiments, the present data suggests a rather nuanced situation in which mutations are neither the directionless force they once were thought to be nor the ruthlessly optimal gradients favoured by engineers. Since selection seems to tune the rate of many genes at once, mutations could at best follow a low-rank approximation of the fitness gradient.
Acknowledgements
Many thanks to Wesley Clawson, Rob Lange, and Inigo Martincorena for comments.
-
According to my partner, who is Swiss, hiking in foggy mountains can be dangerous. So you should actually try to head home rather than up. ↩
-
Perhaps the relative sample complexity of gradient-free vs. gradient-based algorithms could indicate whether evolution’s results would require gradient estimates. ↩
-
In humans and other vertebrates, for example, on average only 1 base pair in every $10^8$ (hundred million) is replicated incorrectly. The replication machinery could therefore copy the famous Kandel textbook (1760 pages, or approximately 2.5 million characters) 40 times over without a mistake! ↩
-
One of the first follow-up studies was the elegant replica plate experiment from Joshua and Esther Lederberg. You can read about it here. ↩
-
Mutation accumulation experiments only work for asexually reproducing organisms. ↩
-
Note, epigenetics is not equivalent to epigenetic inheritance, and is not necessarily environmentally induced either. Epigenetically-determined mutation rates are more likely a by-product of gene expression than a consequence of environmental factors. ↩
-
A reason might be that decreased mutation rates in essential genes have proven advantageous for many millions of years, because their function is highly conserved across species and environments. The importance of non-essential genes, on the other hand, varies on much shorter time scales. Selection might therefore only be able to upregulate mutation rates for genes under strong and recurring positive selection. ↩
-
That’s right, natural selection is not the only cause of evolution. Genetic drift is real, and it limits the efficiency of natural selection. ↩
-
More precisely, the inverse effective population size, which accounts for, e.g., the size of past populations. It is typically much smaller than actual population size; humans have a effective population size of only about 13,000 individuals. This means that genetic drift plays are relatively large role in us compared to, for example, E. coli, which has an effective population size of about 13 million individuals. ↩